
 One of the most iconic observations in biology is Charles Darwin's study of the finches of the Galapagos Islands, and his realization that they had all arisen from the same ancestral bird population. While pigeon breeding (as well as animal husbandry in general) had a long lineage in Europe, there was something about seeing the results of natural (as opposed to human) selection that gave Darwin (at right) a real world foundation for the concept that species evolved over time. A particularly significant observation made by Darwin is that the finches had evolved a variety of beak shapes that permitted them to exploit non-overlapping ecological niches, thereby providing a basis for why evolution had selected these particular varieties of finches and the biological advantages accruing to teach species that helped explain how evolution occurred (i.e., by natural selection).
One of the most iconic observations in biology is Charles Darwin's study of the finches of the Galapagos Islands, and his realization that they had all arisen from the same ancestral bird population. While pigeon breeding (as well as animal husbandry in general) had a long lineage in Europe, there was something about seeing the results of natural (as opposed to human) selection that gave Darwin (at right) a real world foundation for the concept that species evolved over time. A particularly significant observation made by Darwin is that the finches had evolved a variety of beak shapes that permitted them to exploit non-overlapping ecological niches, thereby providing a basis for why evolution had selected these particular varieties of finches and the biological advantages accruing to teach species that helped explain how evolution occurred (i.e., by natural selection).
The evolution of Darwin's finches has been understood as an example of adaptive radiation, where a founder population gives rise to related species having ecological and genetic biodiversity from each other and the founder population. Despite intensive study, the taxonomy of the finches is not completely clear (being based on morphology and breeding patterns). However, factors involved in speciation were thought to include geographic isolation (from S. America), volcanic activity, variations in sea levels due to glaciation, and the effects of climatic variations (for example, from El Nino events).
This week, an international interdisciplinary group of genomic researchers* reported in Nature the genome-wide sequencing of 120 individual finches representative of all the Darwin finch species plus two related species of birds (Tiaris bicolor and Loxigilla noctis) for comparison. (Lamichhaney et al., "Evolution of Darwin's finches and their beaks revealed by genome sequencing," Nature 2015). As reported therein, mitochondrial DNA sequencing confirmed that there are 14 species of finches from the Galapagos Islands and one from the nearby Cocos Islands that have evolved over the past 1.5 million years; advantageously, none of the species has gone extinct (inter alia, through human predation).
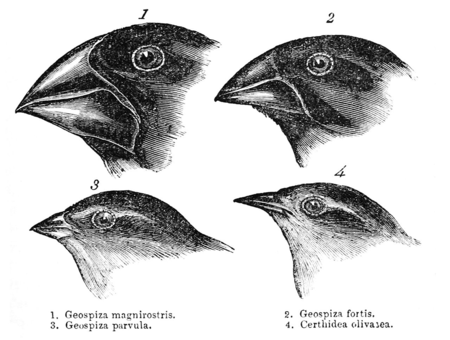
The study comprises ten-fold genome sequence coverage, assembled relative to a female medium ground finch, Geospiza fortis. Effective population sizes of 6,000 – 60,000 for each species were found, and genetic diversity within each species population was found to be similar to other birds and described as being "considerable." Based on the variation found in the sequencing study and using the tools of phylogenetic tree construction supported by these results the study confirmed generally the results of classical taxonomic determinations that the first group to split from the ancestral finch population were the warbler finches (Certhidea sp.) around 900,000 years ago, and that a "rapid radiation" of ground and tree finches were the latest evolutionary split to have occurred from 100,000-300,000 years ago. However, genomic sequencing also found some "interesting deviations" as described in the report:
A network constructed from autosomal genome sequences indicates conflicting signals in the internal branches of ground and tree finches that may reflect incomplete lineage sorting and/or gene flow.
The authors found evidence in introgression between species (interspecies gene flow) that could explain discrepancies between phylogenies based on morphology and genome sequences, wherein they considered hybridization between species to have resulted in mixed ancestry individuals.
The authors further reported the results of genome-wide scans of finch populations that "are closely related but show different beak morphology: G. magnirostris and G. conirostris on Española have blunt beaks, whereas G. conirostris on Genovesa and G. difficilis on Wolf have pointed beaks." These studies used non-overlapping 15-kilobase (kb) windows "to identify regions with the highest fixation indices . . . between groups." Taking the results of the 15 "most significant" genomic regions, the authors reported that they found six contained genes that had been associated with craniofacial development in mammals and beak development in birds; these included calmodulin (CALM), goosecoid homeobox (GSC), retinol dehydrogenase 14 (RDH14), ALX homeobox 1 (ALX1), fibroblast growth factor 10 (FGF10) and forkhead box C1 (FOXC1). Further, beak diversity was found to be associated with a 240 kb haplotype encompassing a gene (ALX1) that encodes a transcription factor involved in craniofacial development:
The region overlaps part of LRRIQ1 (leucine-rich repeats and IQ motif containing 1), the entire ALX1 gene and about 130 kb downstream of ALX1. No previous report indicates that LRRIQ1 has a role during development in vertebrates. By contrast, ALX1 is an excellent candidate for variation in beak morphology. It encodes a paired-type homeodomain protein that plays a crucial role in development of structures derived from craniofacial mesenchyme, the first branchial arch and the limb bud, and on migration of cranial neural crest cells, highly relevant to beak development. Loss of ALX1 in humans causes disruption of early craniofacial development. Also involved in the related species of the medium ground finch (Geospiza fortis).
Specific versions of the genetic haplotype in this 240 kb region were found associated with specific beak morphologies. For example, "[a]ll individuals in the blunt beak category were homozygous for a blunt beak-associated haplotype (denoted B)." Conversely, the pointed beak phenotype was associated with homozygosity for haplotype P "consistent with their phenotypic appearance (sharp-beaked ground finches)." The authors concluded that "[a] phylogenetic tree based on this region revealed a deep divergence between the B and P haplotypes that must have occurred soon after the split between warbler finches and other Darwin's finches." Moreover, "[t]he derived ALX1-B haplotype associated with blunt beaks has a long evolutionary history (hundreds of thousands of years), because its origin predates the radiation of vegetarian, tree and ground finches."
The authors also reported that they found "335 fixed differences between the B and P haplotypes (BB and PP) and that "[t]he short branch lengths among B haplotypes are consistent with a selective sweep" (i.e., a suppression of heterozygosity in the region). With regard to beak phenotype, so-called "derived" alleles (to distinguish later-arising genetic changes from the "ancestral" forms) were found to be "aggregated" in the region of ALX1, and 8 of the 335 conserved differences occurred at conserved sites, where the B haplotype "carried" the derived allele at seven of them.
Looking at these derived alleles, the authors found that four of these were at sites that in the human genome corresponded to transcription factor binding sites, suggesting a mechanism for the observed effects of these alleles on beak phenotype. Two other changes comprised missense mutations in ALX1 itself (L112P and I208V) at highly conserved residues in both bird and human versions of this gene. In the context of sequence comparisons of ALX1 between bird species these changes were considered by the authors to be "non-neutral," further affirming their putative role in adaptive radiation of finch beak morphology.
One particular observation supporting a role for ALX1 in beak diversity came from the results of the genome-wide scan on individuals from G. fortis species, which were found to be polymorphic, which the authors considered "particularly interesting," at least in part because this genetic polymorphism was associated with phenotypic diversity in beak shape in this species seen in field observations. Additional genotyping of 62 G. fortis birds found a diagnostic single nucleotide polymorphism (SNP) that was significantly associated with beak shape (reported as P = 8.8 × 10−5). Phenotypically, PP homozygotes tended to have proportionately long, pointed beaks, BB homozygotes had proportionately deep, blunt beaks, whereas heterozygotes (BP) had intermediate beak shapes.
The authors also found evolutionary change attributable to the past few decades resulting from natural selection on beak size and shape of G. fortis on Daphne Major Island, and that "genetic variation in beak shape has been increased through introgressive hybridization with two species of Geospiza, [Geospiza] scandens and [Geospiza] fuliginosa, that have relatively pointed beaks." Genetic analysis of haplotype frequencies between hybrid birds and backcrosses (to the original type) led these researchers to conclude that "ALX1-P alleles introduced by introgressive hybridization most probably contributed to evolution of more pointed beaks in 1987 following natural selection as a result of a change in food supply in the 1985–86 drought."
The authors concluded that beak shape diversity is "polygenic" based on their association of variations in beak shape with 15 genetic loci, and that "the ALX1 locus contributes to beak diversity, within and among species."
* The authors are from the Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden; the Department of Animal Breeding and Genetics, Swedish University of Agricultural Sciences, Uppsala, Sweden; the Department of Plant Physiology, Umeå University, Umeå, Sweden; the Department of Ecology and Evolutionary Biology, Princeton University, Princeton, New Jersey; and the Department of Veterinary Integrative Biosciences, Texas A&M University, College Station, Texas.