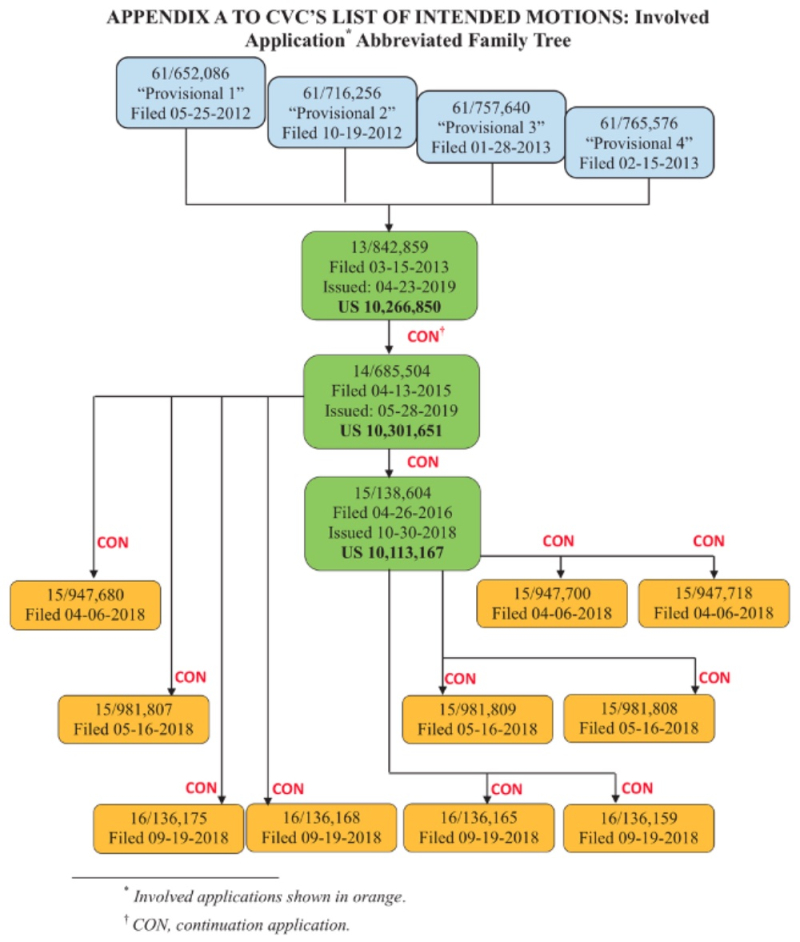
On May 20th, Junior Party the University of California, Berkeley; the University of Vienna; and Emmanuelle Charpentier (collectively, "CVC") filed its Substantive Preliminary Motion No. 1 in Interference No. 106,127 (which names ToolGen as Senior Party), asking the Patent Trial and Appeal Board for benefit of priority to U.S. provisional application No. 61/652,086, filed May 25, 2012 ("P1"), U.S. Provisional Application No. 61/716,256, filed October 19, 2012, ("P2"), and U.S. Provisional Application No. 61/757,640, filed January 28, 2013 ("Provisional 3"), pursuant to 37 C.F.R. §§ 41.121(a)(1)(ii) and 41.208(a)(3) and Standing Order ¶ 208.4.1. The relationships between the patents and applications in the '127 interference are set forth in this chart (filed in CVC's earlier preliminary motion in the '115 Interference):
The significance of the Board granting this motion with regard to the P1 or P2 provisional applications would be that CVC would be Senior Party, with all the presumptions benefits of Senior Party status.
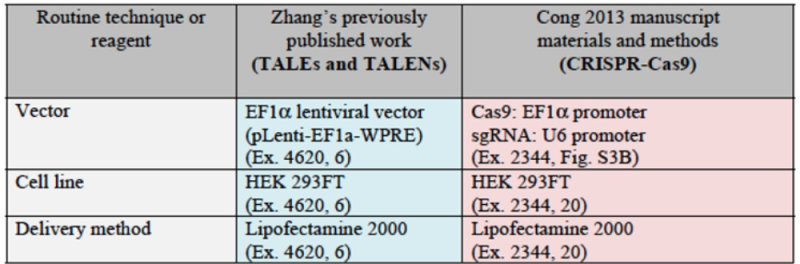
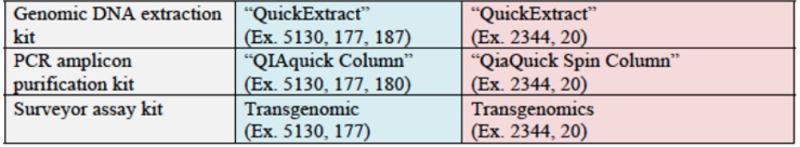
CVC argues that its inventors invented eukaryotic cell CRISPR using a single-molecule guide RNA (sgRNA) that is described in each of the three provisional applications. Once that breakthrough had been achieved, CVC argues that adapting CRISPR to eukaryotic cell environment would have been "pretty straightforward" (quoting Dr. Luciano Marraffini, who informed the Broad inventors of the sgRNA embodiment in June, 2012 (see "CVC Files Motion in Opposition to Broad Priority Motion"). CVC supports this assertion with contemporaneous consistent statements from Rodolphe Barrangou, Erik Sontheimer, Samuel Sternberg, and Dana Carroll, as well as Jennifer Doudna; by the existence of "existing platforms that had already been successfully used with the two incumbent systems: zinc-finger nucleases ("ZFNs") and transcription activator-like effector nucleases ("TALENs")"; and by the successful practice of CRISPR by several groups (including ToolGen) "[j]ust months after CVC presented this work and the absence in the reports from any of these groups of "any 'special' adaptations or conditions needed to achieve CRISPR gene editing in eukaryotic cells." Citing extensively from the record in the '115 Interference, CVC asserts that "investigators from Broad copied CVC's system, applied well-known reagents, cell lines, and vectors, and simply followed the manufacturer's protocols to obtain the results that Broad published in 2013 ('Cong 2013')" and that "CVC itself employed expression vectors and techniques commonly used for ZFNs and TALENs to deliver the sgRNA CRISPR-Cas9 system into eukaryotic cells."
CVC also addresses the PTAB's decision not to grant CVC's patents and applications in the '115 patent the benefit of priority to the P1 and P2 provisional applications sought here; the basis for that decision, according to CVC was that it was made "without the benefit of the now well-developed evidentiary record." Specifically, CVC argues that "[t]he prior decision credited assertions that have been seriously undermined by evidence presented during the priority phase of the '115 interference." That record is cured herein, CVC argues, because the motion "presents new evidence and highlights the specific description in P1 (all of which is carried over to P2) not addressed in the PTAB's prior decision on motions," the focus being on the P1 provisional application because an affirmative decision in this priority document would made CVC the Senior Party. Part of that evidence is that the P1 provisional "contemplates and teaches that the sgRNA CRISPR-Cas9 system can be microinjected as a pre-assembled ribonucleoprotein ("RNP") complex into embryos, including fish cells ("E1"), which obviates the concerns alleged in the '115 interference" (emphasis in brief). Why these aspects of the P1 disclosure are significant, CVC explains, is that the concerns raised by Broad regarding eukaryotic CRISPR embodiments ("RNA and protein expression, co-localization, and assembly") are avoided because the CRISPR-Cas9 complex is already formed in vitro prior to being microinjected into the embryo. RNA degradation would also not have been a concern because it was known that sgRNA was protected from degradation in the complex by the Cas9 protein. Nuclear localization, another putative concern, is avoided by direct microinjection into the nucleus, not would chromatin structure impede CRISPR gene editing activity in embryonic cell because eukaryotic DNA adopts an "open conformation" during cell division in embryos, according to CVC. Finally, the skilled worker would not expect there to be toxicity of CRISPR complex in eukaryotic cells because, according to the brief, microinjection is known to be a "low toxicity" procedure.
It should be noted that these arguments take advantage of the principle that reduction to practice requires only one embodiment falling within the scope of the claims and this argument does not address introduction of CRISPR-Cas9 components into non-embryonic eukaryotic cells. Such embodiments are addressed in the separate argument CVC makes with regard to introduction of CRISPR-Cas9 components into human cells using expression vectors; CVC states that "the alleged concerns have been overstated in [this] context." For example, CVC notes that P1 discloses use of Cas9 from Streptococcus pyogenes, which causes strep throat and thus "flourishes within the same temperature, pH, and ion concentration ranges as most mammalian cells." Also disclosed in P1 is the use of nuclear localization signals to facilitate proper location within a eukaryotic cell.
As to the significance of the Board's earlier decision in the '115 Interference, CVC argues that this decision is non-final (that Interference is on-going and the Board's decisions subject to appeal). In addition, CVC argues that "[t]he PTAB's decision also gave undue weight to certain statements by Doudna, which were misinterpreted as expressing doubt about whether the system would work in eukaryotes." Somewhat cleverly (because those statements have proven to be compelling before the Board both in the '115 Interference and the earlier Interference No. 106,048), CVC argues that they are not relevant evidence, because the extent to which CVC's priority applications disclose at least one operative embodiment within the scope of the Count "is an 'objective' assessment of what is disclosed within the 'four corners' of the specification, viewed "from the perspective of a person of ordinary skill in the art," citing Ariad Pharms., Inc. v. Eli Lilly and Co., 598 F.3d 1336, 1351 (Fed. Cir. 2010) (en banc). To further provide the Board with evidence contrary to these statements, CVC submits a declaration by Inventor Doudna that places the statements in "proper" context. And CVC provides authority for the proposition that an inventor's "acknowledgement of the complexities of the science does not negate the disclosure" in Frazer v. Schlegel, 498 F.3d 1283, 1288-89 (Fed. Cir. 2007), in which an inventor admitted a subjective lack of certainty regarding his invention that the Court held did not "negate or contradict" constructive reduction to practice. Under these circumstances, CVC argues that the Board must "consider the merits anew" in deciding that CVC is entitled to priority benefit to the P1 and P2 provisional applications.
The brief sets forth the "new evidence" presented in support of their motion:
• First, the PTAB's decision on motions in the '115 interference did not address whether direct injection of a pre-assembled RNP complex into an embryo, as contemplated by P1, would trigger the same alleged concerns as embodiments relying on vector expression.
• Second, the PTAB's decision on motions did not address description in P1 regarding: the analogous nature of ZFNs and TALENs; routine uses of nuclear localization signals and codon selection; and the inventors' appreciation of the dynamic nature of chromatin.
• Third, the PTAB's decision on motions did not consider evidence that has since come to light that undermines the initial allegations by Broad that P1 fails to disclose necessary "special" instructions and adaptations for applying CRISPR-Cas9 in eukaryotic cells.
• Fourth, Broad's arguments in the '115 interference relied on a misinterpretation of quotes by Doudna and Carroll. Testimony from Doudna, Carroll, and others with first-hand knowledge (Marraffini, Sontheimer, Barrangou, Sternberg) is provided here, to inform what people in the field thought and the expectation that CRISPR-Cas9 would work in eukaryotes.
The brief then applies each of these new items of evidence to the disclosure of the P1 and P2 provisional applications in making CVC's argument that both priority documents satisfy the disclosure requirements for CVC to be entitled to priority benefit. This argument involves a comparison between eukaryotic CRISPR and successful microinjection of ZFNs and TALENS known in the prior art; that the skilled worker would have considered these systems to be the closest analogous systems to CRISPR-mediated gene editing in eukaryotic cells; that ToolGen itself had admitted the similarities between CRISPR-, ZFN-, and TALEN-mediated gene editing in eukaryotic cells; and that the other systems used by the Board as (negative) comparators in the '115 Interference (such as Group II introns, ribozymes, and riboswitches) were not apposite.
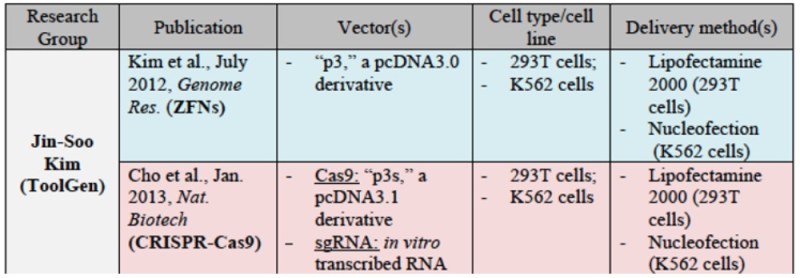
Moreover, CVC asserts once again the evidence that, once sgRNA was disclosed by Doudna et al., several (six) groups used it to achieve successful CRISPR-mediated gene editing in eukaryotes. The brief sets out relevant details of each of these groups' successes: for Broad:
for ToolGen:
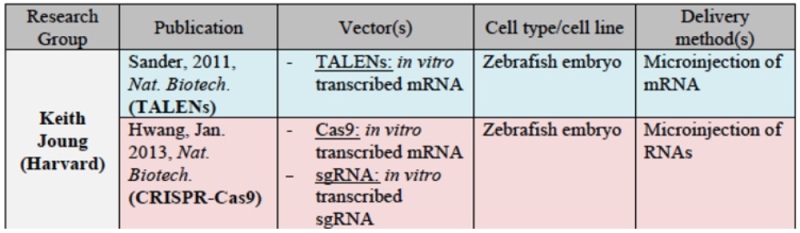
for Harvard:
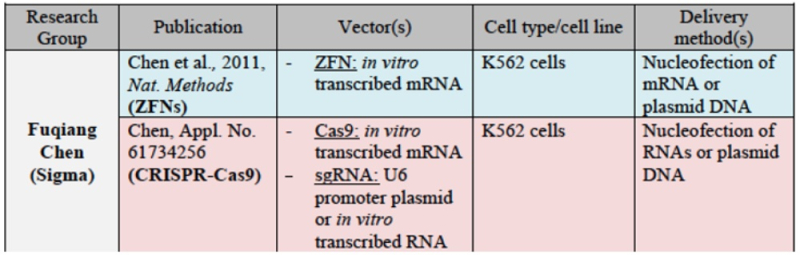
and for Sigma:
The brief also sets forth in detail the correspondence between the elements of the Count in the Interference and the disclosure in the P1 (and P2) references for each of these embodiments (E1, E2, and E3):
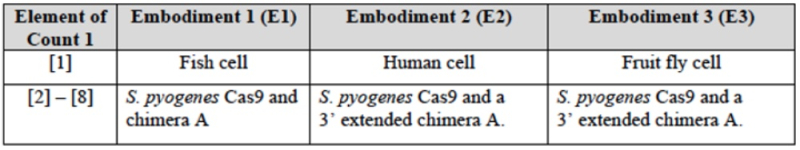
wherein CVC asserts (as it did in the '115 Interference; see "Berkeley Files Substantive Motion No. 2 to be Accorded Benefit to Earlier Priority Application in Interference") the three embodiments CVC contends fall within the scope of the Count (designated E1 for introduction into fish embryos; E2 for introduction into human cells; and E3 for introduction into fruit fly cells) and that the P1 (and P2) provisional applications thus disclose constructive reduction to practice of CRISPR-Cas9 gene editing in eukaryotic cells. CVC also sets forth examples of third parties using similar embodiments of their CRISPR technology published after the filing date (May 25, 2012) of the P1 provisional application:
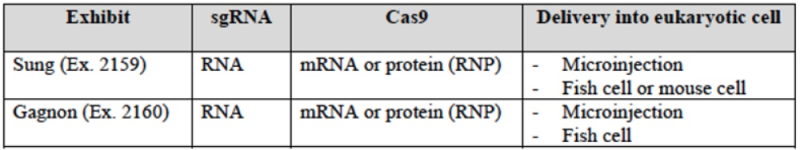
The brief also addresses the "concerns alleged in the '115 interference" and attempts to allay them for the Board, enumerating RNA degradation; co-localization of the components of the CRISPR-Cas9 complex; cellular conditions; nuclear localization; codon optimization; the effects of chromatin structure and toxicity, none of which are relevant, as CVC argues, for eukaryotic CRISPR-Cas9 embodiments using microinjection of in vitro assembled CRISPR-Cas9 complexes.
Similar explication and explanation, particularly of the E2 embodiment introducing CRISPR-Cas9 into human cells using an expression vector are set forth in CVC's brief; in this regard the brief similarly addresses the "concerns" asserted against the P1 and P2 provisional applications in the '115 Interference, while at the same time discounting these concerns as "(i) . . . not [being] recited elements of the count and (ii) . . . not [being] required for practicing an embodiment within the scope of the count." Much of these arguments rely on what the person of ordinary skill in the art would have understood at the time, such as using strong promoters to produce effective amounts of the sgRNA and Cas9 protein; using NLSs to increase nuclear localization likelihood; using S. pyogenes-derived Cas9 that purportedly "flourish" under eukaryotic cellular conditions; and that the P1 (and P2) disclosure recognize that chromatin structure is sufficiently "dynamic" for the CRISPR-Cas9 complex to gain access to portions of the genome targeted by the sgRNA for CRISPR-mediated gene editing to be achieved.
CVC argues in the alternative that should the Board find wanting its disclosure evidence in the P1 provisional application, the P2 application contains additional disclosure that satisfies the requirements for constructive reduction to practice of at least one embodiment of the invention falling within the scope of the interference Count.
In a separate section, prefaced by the assertion that it is "not required," CVC sets forth statements contemporaneous with the P1 and P2 provisional application filing dates that eukaryotic embodiments of CRISPR using sgRNA were expected to be operative in eukaryotic cells. These statements were made, according to CVC, by Luciano Marraffini; Erik Sontheimer; Rodolphe Barrangou (wherein each of the foregoing were present and heard Jennifer Doudna's disclosure of sgRNA CRISPR embodiments at the Fifth Annual CRISPR Research Conference held at Berkeley in June of 2012); Dana Carroll, editor of a review article on CRISPR; Samuel Sternberg (a graduate student in the Doudna laboratory); and Jennifer Doudna herself (offering the "context" CVC intends to use to rebut Broad's negative interpretations of her statements regarding prospects for eukaryotic CRISPR embodiments).
Finally, CVC sets out the continuous priority chain from the P1 provisional application through the applications and patents at issue in this interference, as set forth in the diagram above.