
Human evolution, once its occurrence was recognized over a century and a half ago, has long been a source of confusion, concern, and controversy (as well as fascination and wonder). The recent explosion in our understanding of the human genome, and particularly genomes of humanity's ancestors, has refined the concept of what it means to be human. For example, it has become common knowledge (thanks to 23andMe, Helix, and others) that modern humans contain small amounts (~2%) of DNA in common with Neanderthals, and that other forms, such as the Denisovans and Homo florensiensis, existed in historical time (i.e., tens of thousands rather than millions of years ago). See D. Reich, "Who We Are and How We Got Here: Ancient DNA and the New Science of the Human Past" for a more detailed description of these genomic relationships between branches of the human family tree.
The genomic fossil record has revealed that Neanderthals and humans had two periods of extensive interbreeding in Europe and East Asia (after their divergence 500,000 to 800,000 years ago): one that occurred ~100,000 years ago and another that happened ~50,000 years ago. As evidenced by the small amount of heterologous (i.e., from the other subspecies) DNA maintained in modern humans, it has been postulated that "purifying selection" against maintenance of the bulk of this DNA was responsible for its elimination. But the persistence of even the small amount of Neanderthal DNA remaining in modern humans raises the question of why this DNA was retained and if there was any functional (i.e., selected-for) component (as opposed to merely random persistence).
Recently, researchers at the University of Arizona* and Stanford** studied the patterns of introgression (where in the genome) of human and Neanderthal DNA in samples from each human subspecies, in an article entitled, "Evidence that RNA Viruses Drove Introgression between Neanderthal and Modern Humans" in the journal Cell. Their results showed enrichment of heterologous "insertion sequence (IS)" DNA at sites that encode virus-interacting proteins (VIPs), suggesting that human-Neanderthal interbreeding was associated with challenges for each species by novel viruses, and that the persistent DNA was somehow advantageous to the "host" species in overcoming or adapting to infection by such xenoviruses. As stated in the paper, "[i]n line with the overall enrichment of Neanderthal ancestry at VIPs being due to adaptive introgression, we found a very strong excess of adaptive IS at VIPs compared to non-VIPs."
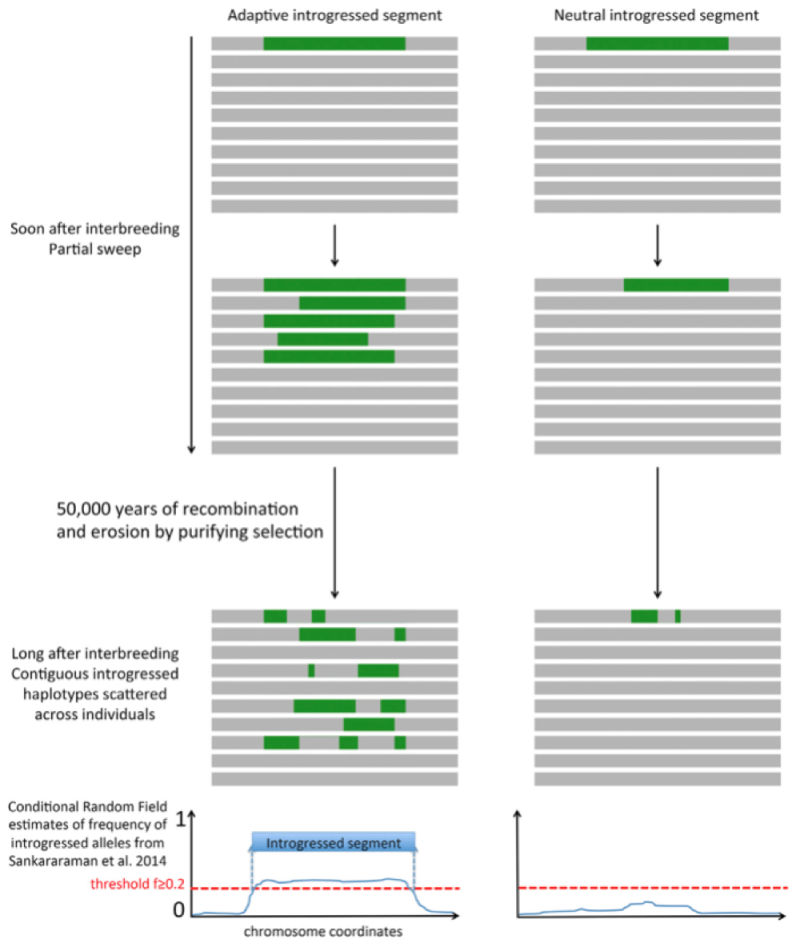 The data presented showed 121 introgressed Neanderthal DNA segments longer than 100 kb that overlapped VIPs sequences in human samples from East Asia (compared with 66 such sequences expected by genomic distribution and probability models) and 103 such sequences (versus 68 expected) in samples from humans of European lineage. This pattern is also observed for introgressions that are more likely to be adaptive (longer and at higher frequency), albeit occurring less often but with even higher relative enrichment levels (36 versus 11 expected segments in East Asian-derived DNA, and 19 versus 6 expected in European-derived DNA). The authors concluded from these results that "out of all long and high-frequency IS from Neanderthals to modern humans, 15% to 32% (54 of 171) in East Asians and 12% to 25% (27 of 105) in Europeans have been positively selected in response to viruses" and that "[i]n total there are 171 and 105 long and high-frequency IS overlapping genes in East Asians and Europeans, respectively." These researchers also tested for introgression of human DNA into the Neanderthal genome, using DNA from the single individual Neanderthal from the Altai Mountains, with similar (but less robust) outcomes.
The data presented showed 121 introgressed Neanderthal DNA segments longer than 100 kb that overlapped VIPs sequences in human samples from East Asia (compared with 66 such sequences expected by genomic distribution and probability models) and 103 such sequences (versus 68 expected) in samples from humans of European lineage. This pattern is also observed for introgressions that are more likely to be adaptive (longer and at higher frequency), albeit occurring less often but with even higher relative enrichment levels (36 versus 11 expected segments in East Asian-derived DNA, and 19 versus 6 expected in European-derived DNA). The authors concluded from these results that "out of all long and high-frequency IS from Neanderthals to modern humans, 15% to 32% (54 of 171) in East Asians and 12% to 25% (27 of 105) in Europeans have been positively selected in response to viruses" and that "[i]n total there are 171 and 105 long and high-frequency IS overlapping genes in East Asians and Europeans, respectively." These researchers also tested for introgression of human DNA into the Neanderthal genome, using DNA from the single individual Neanderthal from the Altai Mountains, with similar (but less robust) outcomes.
With regard to which viruses might have been responsible for the persistence of these introgression events, the researchers used 20 modern human viruses known to interact with ten or more VIPs, as proxies for the ancient related viruses that infected humans at the time of interbreeding, and ones that were "evenly distributed between RNA viruses (2,684 VIPs, of which 1,563 were specific for RNA viruses) and DNA viruses (2,547 VIPs, of which 1,426 were specific for DNA viruses)." Their results showed no preference for RNA versus DNA virus VIPs from human DNA of East Asian origin, but did detect a "strong bias" for RNA virus VIPs from European-derived DNA in long, high-frequency IS. These results were even more striking when the researchers excluded VIP genes known to interact with other microbial species (bacteria, Plasmodium) and human "immune genes." Analyzing these RNA virus VIP genes more closely, the authors report that VIPs that interact with lentivirus (HIV), orthomyxovirus (influenza A) and flavivirus (hepatitis C) had ("by far") the highest number of VIPs associated with Neanderthal IS in the human genome. (The authors are careful to note, however, that "[t]he enrichments of VIPs in IS represent statistical associations, and more evidence is required to demonstrate causality.")
Delving even further into these interactions regarding the functions of the VIP genes associated with introgression, these researchers found that "a crucial early step of infection, ''virion attachment to host cell,'' was among the most strongly over-represented GO [Gene Ontology] functions among all VIPs, both in Europe (5 VIPs instead of 0.8 expected by chance[]), and in East Asia (4 VIPs versus 0.75 expected by chance[]." On the other hand, those VIPs known to be involved in RNA virus attachment were all known to be involved with lentiviruses (ICAM1, CD209/DCSIGN, HSP90AB1, and CLEC4M), wherein CD209/DCSIGN, HSP90AB1, and CLEC4M interact with HVC; there was one DNA virus attachment VIP (PVRL2) detected. Also reported were detection of VIPs involved in ''viral genome replication,'' which also showed strong over-representation in human DNA from European sources (17 VIPs versus 7.8 expected) and those known to interact with lentiviruses (17 LT-VIPs versus 6.9 expected). They also report ("[i]ntriguingly") that "a large number of these 'viral genome replication' VIPs were again VIPs that interact with HCV as in the case for virion attachment (LTF, EIF2AK2/PKR, ATG5, MAVS, CD209/DCSIGN, CLEC4M, VAPB; all HCV VIPs: 7 versus 1.7 viral genome replication VIPs, []; HCV LT-VIPs: 7 viral genome replication VIPs versus 1.5 expected)."
A part of the conclusions these authors draw from these data is that they are consistent with the ''poison-antidote'' hypothesis, which suggests that "the interactions between modern humans and Neanderthals exposed each species to novel viruses while gene flow between the species afforded a measure of resistance by allowing VIPs that were already adapted to the presence of specific viruses in the donor species to cross species boundaries and provide adaptive function in the recipient species" (although they also caution that this interpretation is only "preliminary" and that they provide "only statistical association"), illustrated below:
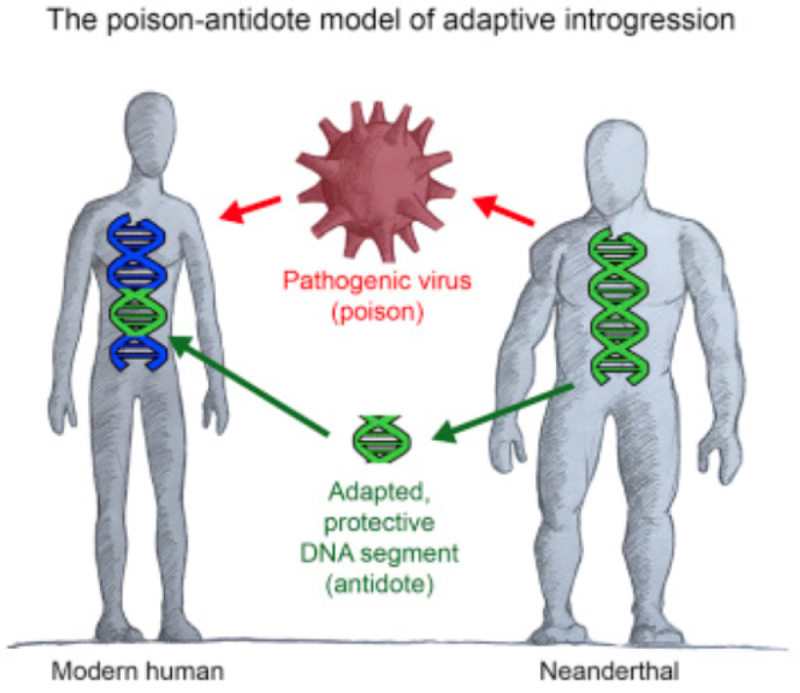
And they conclude with the following statement of the significance of their results (preliminary or not):
[T]hese results suggest that the genomes of humans and other species contain signatures of past arms races with diverse viruses and other pathogens, making it possible to use host genomic signatures to study ancient interactions with ever present and ever shifting viral and other pathogens. In this respect, it is worth noting that even though we focused on Neanderthal ancestry, we anticipate that it should also be possible to study the impact of ancient epidemics on introgression from Denisovans to modern humans, especially in populations such as Melanesians with a larger percentage of Denisovan ancestry[]. The results of such studies should provide important insights into the dynamics of past, present, and future epidemics [citation omitted].
*D. Enard **D. Petrov