
One of the consequences of the explication of human genomic DNA by the Human Genome Project and related efforts has been a better understanding of anthropological history, i.e., how the human population has changed geographically over time. This better understanding has been set forth in broad strokes based on mitochondrial DNA differences in historical and contemporary human populations (leading to the "African Eve" hypothesis of human ancestry) as well as analyses of Y-chromosome polymorphism (or lack of it; Genghis Khan has a frightfully large number of descendants). References to this part of the human story are many and include Deep Ancestry by Spencer Wells (National Geographic: 2006), and The Seven Daughters of Eve: The Science that Reveals our Genetic Ancestry by Brian Sykes (Carrol & Graf: 2002).
Recently a group of researchers from the U.S., U.K and Australia have conducted another type of study, looking at the fine structure of human population diversity in England. These results were reported in Nature in an article entitled "The fine-scale genetic structure of the British Population," Nature 519: 304-314 (19 March 2015); doi:10.1038/nature14230. These authors* showed what the authors termed "a rich and detailed pattern of genetic differentiation with remarkable concordance between genetic clusters and geography," based on genome-wide single nucleotide polymorphism (SNP) analyses of "a carefully chosen geographically diverse sample of 2,039 individuals." The results include maps of where Anglo-Saxon influence is strongest (and where it is absent) and reveal evidence of migrations from the European continent that predates the Roman conquest. The authors also report that the non-Saxon regions of Britain were populated by "genetically differentiated subgroups" of people of Celtic ancestry possessing greater genetic diversity than heretofore appreciates.
The populations studied were from throughout Great Britain (as conventionally defined to include England, Scotland, Wales and Northern Ireland) and comprised a data set termed the People of the British Isles (PoBI). By using samples from rural areas wherein all four grandparents of living study participants livd within 80 km of each other, the authors assert that they had "effectively sample[d] DNA from the grandparents" (who had an average date of birth in 1885). The study also interrogated the same SNP patterns from 6,209 samples from ten continental European countries from 500,000 SNPs.
The authors reported that "[t]he correspondence between the genetic clusters and geography [was] striking: most of the genetic clusters are highly localized, with many occupying non-overlapping regions." As set forth in the Figure, 17 clusters were chosen for convenience of the analysis. The clustering and their integrity is shown for Cornwall and Devon, where the genetic clusters "closely match" existing county boundaries and (perhaps less surprisingly) in Orkney, the region off the northern coast of Scotland, reflecting the contributions of Norse Viking "settlers."
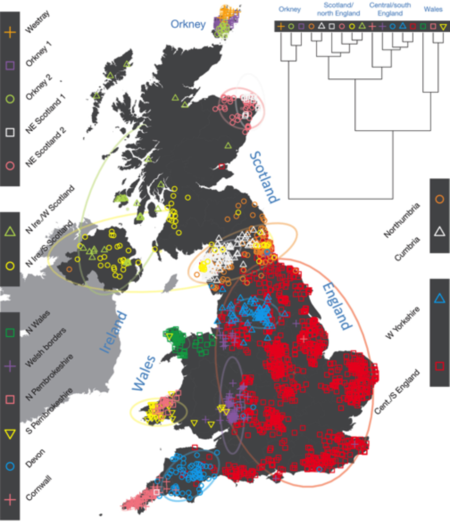
The article also supplies a "tree" reflecting the relationships between the clusters, which the authors describe as follows:
The coarsest level of genetic differentiation (that is, the assignment into two clusters) separates the samples in Orkney from all others. Next the Welsh samples separate from the other non-Orkney samples. Subsequent splits reveal more subtle differentiation (reflected in the shorter distances between branches), including separation of north and south Wales, then separation of the north of England, Scotland and Northern Ireland from the rest of England, and separation of samples in Cornwall from the large English cluster. There is a single large cluster (red squares) that covers most of central and southern England and extends up the east coast. Notably, . . ., this cluster remain[ed] largely intact and contains almost half the individuals (1,006) in [the] study.
The authors correctly point out that the genetic differentiation they detect is small and the differentiation between clusters is "subtle." When compared with existing European populations the genetic results showed ancestry in all clusters from western Germany, Flemish Belgium, northwestern France, Denmark, southern France and Spain. Norwegian ancestry was found disproportionately in the Orkney-derived clusters ("and to a lesser extent the clusters involving Scotland and Northern Ireland") and Swedish groups were found in the ancestry profiles of other British clusters.
The authors conclude that:
The clustering [shown inter alia in Fig. 1] is notable both for its exquisite differentiation over small distances and the stability of some clusters over very large distances. Genetic differentiation within the UK is not related in a simple way to geographical distance. Examples of fine-scale differentiation include the separation of: islands within Orkney; Devon from Cornwall; and the Welsh/English borders from surrounding areas. The edges between clusters follow natural geographical boundaries in some instances, for example, between Devon and Cornwall (boundaries the Tamar Estuary and Bodmin Moor), and Orkney is separated by sea from Scotland. However, in many instances clusters span geographic boundaries; for example, the clusters in Northern Ireland span the sea to Scotland.
There conclusions support a reconstruction about how migrations produced the modern British distribution:
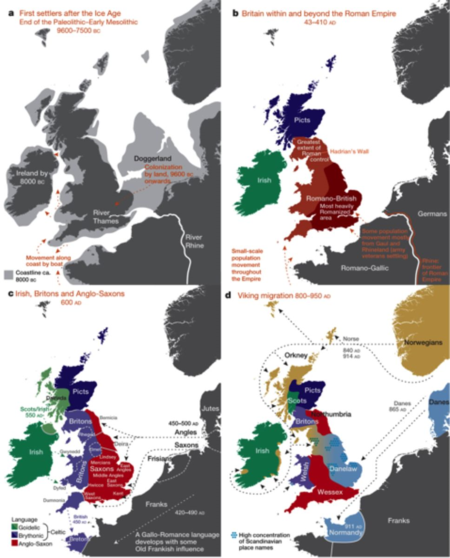
The authors were able to interpret these results to give a picture about how migrations of various groups has changed over time:
[T]he earliest migrations whose descendants survive to make a substantial contribution to the present population are best captured by three groups in our European analyses, GER6 (western Germany), BEL11 (Belgium), and FRA14 (north-western France). These groups still contribute to current patterns of population differentiation []. Other European groups may reflect early migrations into the UK, but with smaller contribution, including SFS31 (southern France/Spain), at least part of DEN18 (Denmark), and possibly parts of Norway and Sweden. A subsequent migration, best captured by FRA17 (France), contributed a substantial amount of ancestry to the UK outside Wales. []Migrations represented by FRA12 essentially only affect Wales and Northern Ireland and/or Scotland. We also see clear signals of some of the known historical migrations and settlements, including the Saxons (GER3, northern Germany, and probably much of DEN18, Denmark) and the Norse Vikings (NOR53–NOR90).
Finally, these researchers applied more sophisticated genetic analytical techniques (related to the rate of decay of shared haplotype segments) to assess population admixture, and found "strong evidence (P < 0.01) that the largest Orkney cluster (Orkney 1) was influenced by a recent (about 29 generations ago, or in 1100 A.D.) admixture event with an overall contribution of ~25% of the DNA from groups in Norway."
The article also notes that the known historical migration events frequently have less of a genetic impact that might be presumed; for example, the authors reported seeing "no clear genetic evidence of the Danish Viking occupation and control of a large part of England" and "no evidence of a general 'Celtic' population in non-Saxon parts of the UK." Rather, with regard to the Celtic populations in Britain the article reports "there were many distinct genetic clusters in these [non-Saxon] regions, and the Welsh clusters "represent populations that are more similar to the early post-Ice-Age settlers of Britain than those from elsewhere in the UK."
The article closes by reminding readers that such genetic studies "can augment archaeological, linguistic and historical approaches to understanding population history" and also "complements them, in providing evidence relating to the bulk of ordinary people rather than the successful elite."
* Stephen Leslie (Murdoch Childrens Research Institute, Royal Children's Hospital, University of Melbourne, Department of Mathematics and Statistics, Victoria, Australia and University of Oxford, Department of Oncology, Oxford UK), Bruce Winney (University of Oxford, Department of Oncology, Oxford UK), Garrett Hellenthal (University College London Genetics Institute, London UK), Dan Davison (Counsyl, South San Francisco, California), Abdelhamid Boumertit (University of Oxford, Department of Oncology, Oxford UK), Tammy Day (University of Oxford, Department of Oncology, Oxford UK), Katarzyna Hutnik (University of Oxford, Department of Oncology, Oxford UK), Ellen C. Royrvik (University of Oxford, Department of Oncology, Oxford UK), Barry Cunliffe (University of Oxford, Institute of Archaeology, Oxford), Wellcome Trust Case Control Consortium 2, International Multiple Sclerosis Genetics Consortium, Daniel J. Lawson (University of Bristol, Department of Mathematics, Bristol UK), Daniel Falush (College of Medicine, Swansea University, Swansea UK), Colin Freeman (The Wellcome Trust Centre for Human Genetics, Oxford UK), Matti Pirinen (University of Helsinki, Helsinki, Finland), Simon Myers (University of Oxford, Department of Statistics, Oxford UK), Mark Robinson (University of Oxford, University Museum of Natural History, Oxford UK), Peter Donnelly (University of Oxford, Department of Statistics, Oxford UK) & Walter Bodmer (University of Oxford, Department of Oncology, Oxford UK)