Cephalopods are fascinating creatures, and their primary living representative -- the octopus -- has recently been the subject of the Academy Award winning documentary "My Octopus Teacher." They are clearly intelligent (as set forth in Peter Godfrey-Smith's Other Minds: The Octopus, the Sea, and the Deep Origins of Consciousness), but it is an intelligence so different from human intelligence that it was an inspiration for the alien first-contact film, Arrival.
Outside the coleoid group, the only living relative are animals of the genus Nautilus, the sole surviving externally shelled cephalopod since the Paleozoic (541 to 252 million years ago). That shell is itself iconic, as both objet d'art and a living illustration of the Fibonacci series in the ratios of the whorls from which the shell is made.
Recently, an international group of researchers (predominantly working in China)* published a paper entitled "The genome of Nautilus pompilius illuminates eye evolution and biomineralization," in the July edition of Nature Ecology & Evolution, elucidating the Nautilus genome. (The octopus genome has been determined previously; see "Octopus Genome Sequenced".) In stark contrast with the octopus genome, the Nautilus genome is parsimonious, the authors characterizing it as "compact, minimalist" with few genes and "slow evolutionary rates in both non-coding and coding regions." Genetic innovations in Nautilus involve gene loss, with expansion and contraction independently of certain gene families resulting in the "pinhole" eye which functions without either cornea or lens. These distinctions with their coleoid cephalopod cousins provide the possibility that genomic comparisons could provide insights into how evolution shaped both cephaloid types, according to the authors.
These researchers report that the Nautilus genome comprises 731 megabasepairs, which is the smallest of the cephalopods, being 2.5-7 fold smaller than their extant coleoid relatives. One feature the authors called "strikingly different" in Nautilus DNA is found in transposable elements (TE), known to be "the driving force in shaping genomic architecture and evolution." TEs comprise 31% of the genome, with retrotransposons, including LINE (long interspersed nuclear element) SINE (short interspersed nuclear element), and LTR (long terminal repeats) elements constituting only a minor portion (6.5%); in contrast, these elements are "a prominent presence" in the coleoid cephalopod genome. Moreover, further analysis indicated that an ancient DNA transposon "burst" occurred once in the Nautilus lineage and that there has been no further expansion of TEs in the Nautilus genome (coleoid cephalopods have had more evolutionarily recent instances of such bursts). Nautilus shares this characteristic with other mollusks, which the authors say suggest slow evolutionary rates in non-coding regions of the Nautilus genome.
Genetic analyses identified 17,710 protein-coding sequences in the Nautilus genome, which is only 53-61% of the number of putative gene sequences found in octopus and squid genomes. On the gene family level this is reflected by "a huge contraction of orthologous gene families" in the Nautilus genome (204 gene families contracted versus only 9 gene families that have expanded). The authors note that 18 domains of centromere protein B were specifically expanded in Nautilus compared with other cephalopods; this protein is thought to "play[] crucial roles in host genome integrity and replication fidelity through the repression of retrotransposons and centromere formation in yeast or humans" consistent with the frequencies of TE sequences found in the Nautilus genome.
On a more global level, comparisons of the Nautilus genome to related (and not-so-related) species was used to assess phylogenetic relationships, using 423 single-copy orthologues from 16 animal genomes as shown in the figure:
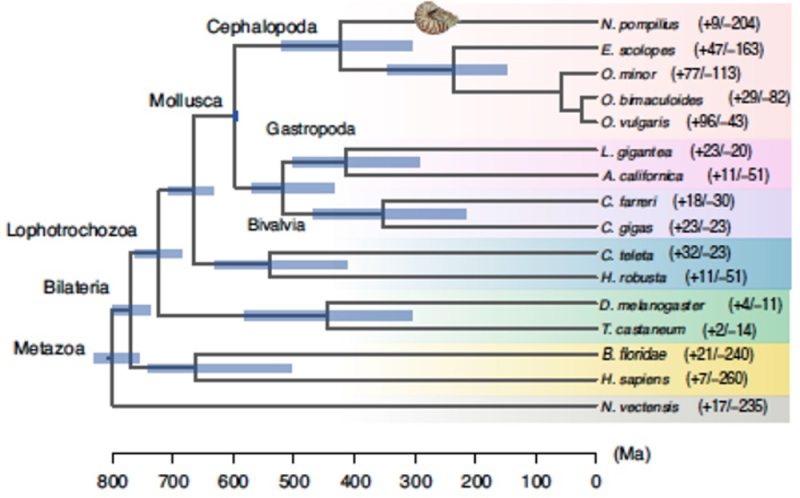
Divergence from the coleoid cephalopods was estimated by these studies to have occurred around the Silurian-Devonian boundary at about 423 million years ago (Ma), consistent with other estimates from geological evidence and "molecular clock" inferences. On a finer scale, these researchers report that N. pompilius populations expanded about 23 Ma ("at the turn of the Miocene") in a "stepwise manner," which expansion then stopped as a result of climate changes (glaciation) at the Mid-Pleistocene Transition (around 1 Ma), and finally fell precipitously around 0.4 Ma during another intense glaciation period termed the Mid-Brunhes Event. These authors conclude that this pattern indicates N. pompilius to be sensitive to "extreme environmental fluctuations" (although it is not evident that they are particularly more sensitive than other aquatic species). Paradoxically octopus species and certain bony fishes thrived during this period according to the paper, suggesting "ecological competition" (i.e.,"nature red in tooth and claw") had much to do with the population record of N. pompilius during these times.
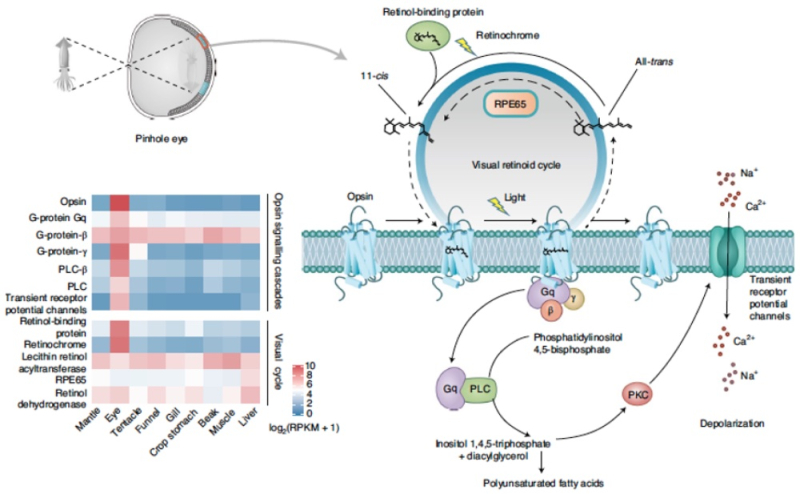
Turning to the pinhole eye, these researchers report the presence of "nearly all" of the genes encoding the core regulatory complex of transcription factors thought to govern lens formation in coleoid cephalopods (PAX6, SIX3/6 and SOX2, among others) in Nautilus, consistent with lens development in the early Cambrian era (541-485 Ma). In Nautilus, an important regulatory gene for lens production (in vertebrates as well as coleoid cephalopods), Nrl/Maf, is missing and genes encoding lens-specific crystallin genes are "contracted" compared to other mollusks like octopus that have lens-equipped eyes. Other specific crystallin genes found expanded in coleoids are missing in the Nautilus genome. Nautilus is known for behaviors (such as coming towards the surface to feed under lower light conditions) indicating visual perception, and were found by these researchers to possess one photoreceptive r-opsin gene and one retinochrome gene, which these authors assert represents "the minimal opsin gene number among known metazoans" (albeit conceding that N. pompilius is almost certainly colorblind). The authors note that light intensity is a more critical faculty for animals like N. pompilius that migrate vertically between depths at sea, and that such sensitivity depends on the 11-cis retinal chromophore which isomerizes in response to light. The N. pompilius genome contains a retinochrome encoding such an isomerase, and in addition these researchers identified an ortholog of a gene family (ten genes) named retinal pigment epithelium-specific protein 65 kDa present in vertebrates that is also found in the Nautilus genome. These genes contain a conserved iron binding site, an "active cavity" site, and a "hydrophobic tunnel" consistent with catalytic activity. The significance of these finding is explained by the authors as:
From a perspective of evolutionary adaptation, the appearance of the pinhole eye is one adaptive breakthrough essential to the nautilus lifestyle of vertical depth migrations, allowing the organism to acquire spatial vision and rapidly cope with hydrostatic pressure within the eye through opening the pupil to seawater. Overall, multiple genomic innovations including gene losses, independent contraction and expansion of specific gene families and presence of associated regulatory networks seem to work in unison to drive the evolution of the pinhole eye in nautilus.
The paper provides a schematic diagram of the Nautilus pinhole eye to put these genetic analyses in context:
Turning to the other characteristic morphological feature of Nautilus, its shell, the authors note that it is made up of aragonite (an orthorhombic crystal of calcium carbonate, CaCO3) and shell matrix proteins (SMP) used as a substrate. The authors report finding genes encoding a total of 78 SMPs with expression pattern concentrated in the shell mantle (72%). Genetic comparisons revealed that 21 Nautilus SMPs shared sequence similarity with analogous genes from bivalves and gastropods and found several conserved domains in these protein across mollusk species, including "laminin, chitin-binding and carbonic anhydrase domains." The authors posit these regions as part of a "core biomineralization toolkit" conserved across external shell-expressing mollusks. On the other hand, 52 of the 78 SMPs encoded in the Nautilus genome were either new or Nautilus-specific, and these researchers speculate that most of the unique SMPs evolved independently in different mollusk species including Nautilus. Indeed, there is one important SMP, Nautilin-63, that these researchers found showed low sequence similarity even within the Nautilus genus. And the top ten SMPs enriched in the Nautilus mantle contain an evolutionarily novel poly(Gly or Gly-Ala) repetitive motif; also detected encoded in these proteins were several repetitive low-complexity domains (RLCDs), in common with other shelled mollusks, that the inventors suggest "could be a unifying principle for molluscan biomineralization, especially for nacre formation."
Finally, the paper discusses the Nautilus immune system, which contains a "highly complex yet comprehensive innate immune components" enabling Toll-like receptor and tumor necrosis factor receptor signaling that regulates "apoptosis, inflammation and immune defences." Also detected were genes encoding IL17R, H-lectin, and IL1, which the authors assert "supports the assumption that nautilus has preserved a more complete repertoire of immune molecules than other cephalopods." An assessment of gene numbers for immune system-specific genes showed expansion of C-type lectin genes (81) compared to coleoid species (wherein 12-33 genes are encoded) and some of these genes have been independently duplicated. Also found to be specifically expanded are interferon-inducible GTPases, genes also known to be implicated in immune function. These genetic features of the immune system-involved genes in Nautilus (and comparisons with coleoid species) is illustrated in this figure:
The authors conclude with this synopsis of their findings:
Genomic evidence reveals that nautilus has undergone lineage-specific innovations in both body plan and behaviour since the Cambrian and retained these extraordinary features after a long evolutionary history. In particular, vertical depth migration in Nautilus and other chambered cephalopods is one of several critical and common strategies needed to avoid predators and budget energy; these may have helped the survival of these species ever since. The emergence of the pinhole eye is a great innovation for switching from directional to spatial vision and rapidly change hydrostatic pressure, making vertical depth migration possible.
Our findings highlight that co-evolutionary loss of core regulatory transcription factors may have driven the evolution of the pinhole eye. Moreover, our proteomic and transcriptomic data suggest that an ancient "core biomineralization toolkit" and new RLCDs coordinately directed the construction of the chamber shell, which has evolved into the buoyancy apparatus needed to adapt to a critical life mode. Taken together, the draft genome of N. pompilius together with multiomics provide a valuable insight into not only the adaptive innovations of the ancestor of cephalopods but also the dynamic evolution of coleoids.
* Key Laboratory of Tropical Marine Bio-resources and Ecology and Guangdong Provincial Key Laboratory of Applied Marine Biology, South China Sea Institute of Oceanology, Chinese Academy of Sciences, Guangzhou; Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences, Guangzhou; Southern Marine Science and Engineering Guangdong Laboratory, Guangzhou; MOE Key Laboratory for Membraneless Organelles and Cellular Dynamics, Hefei National Laboratory for Physical Sciences at the Microscale, CAS Key Laboratory of Brain Function and Disease, School of Life Sciences, Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei; Zhejiang Key Laboratory of Aquatic Germplasm Resources, College of Biological and Environmental Sciences, Zhejiang Wanli University, Ningbo; Biomarker Technologies Corporation, Beijing; State Key Laboratory of Biocontrol, College of Life Sciences, Sun Yat-sen University, Guangzhou; Department of Neuroscience and Developmental Biology, University of Vienna.