In its turn, Junior Party The University of California/Berkeley, the University of Vienna, and Emmanuelle Charpentier (collectively, "CVC") filed its motion in opposition to Senior Party The Broad Institute, Harvard University, and the Massachusetts Institute of Technology (collectively, "Broad") motion for priority in Interference No. 106,115. CVC's motion challenges Broad's priority claim and the bases Broad set forth therein, rebutting Broad's legal arguments and mentioning more than once that Jennifer Doudna and Emmanuelle Charpentier received the 2020 Nobel Prize in Chemistry (and by implication that the Broad inventors had not).
CVC's motion is based on two principles. First, as supported by deposition testimony of Dr. Luciano Marraffini (compelled by the Board's grant of CVC's motion to require compliance), CVC argues that Broad inventor Zhang derived the invention claimed in the patents-in-interference from disclosure of CVC's conception from Dr. Marraffini. Dr. Marraffini was in possession of CVC's invention because he was a confidential reviewer of the manuscript later published in Science as Jinek et al. (2012, "A Programmable Dual-RNA-Guided DNA Endonuclease in Adaptive Bacterial Immunity," Science 337: 816–21). Dr. Marraffini also attended a CRISPR conference at Berkley on June 26, 2012 where the Doudna lab disclosed its CRISPR findings. Indeed, the brief contains a comparison between what was disclosed at the meeting, what Dr. Marraffini disclosed to Dr. Zhang, and what Dr. Zhang communicated to his colleague Dr. Cong, first author on the paper published in the January 2013 issue of Science and containing Broad's disclosure of CRISPR practiced in eukaryotic cell:
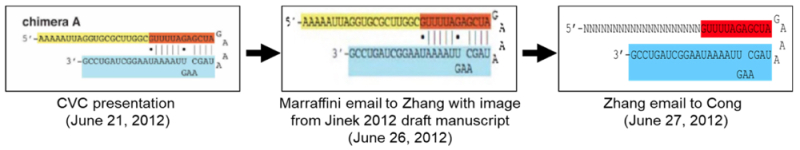
CVC sets out testimony from Dr. Marraffini that he immediately communicated the key finding disclosed at that meeting by the Doudna/Charpentier group: that CRISPR could be performed by single-guide RNA (sgRNA) comprising tracr and sequence-specific mature crRNA RNA fragments covalently linked in association with the Cas9 protein. Prior to this discovery CVC relies on Dr. Marraffini's testimony that Dr. Zhang) understood that formation of functional CRISPR complexes required RNAse III cleavage of unprocessed tracr and crRNA.
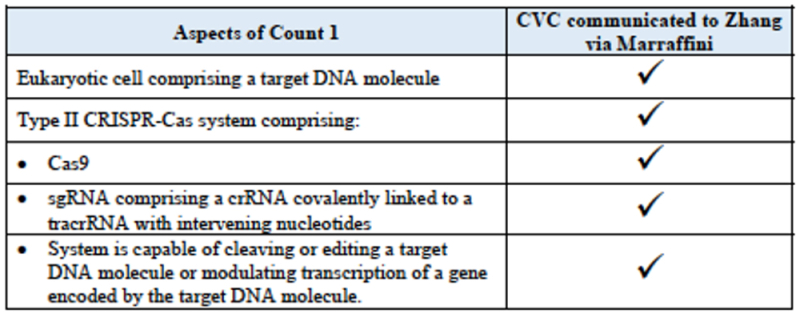
Second, CVC argues that once its seminal finding was made public, that CRISPR could be performed using sgRNA, no fewer than five labs ("Church (Harvard), Kim (ToolGen), Joung collaboration, and Chen (Sigma-Aldrich)"), including the Zhang lab, were able to perform eukaryotic CRISPR using routine, conventional laboratory methods, as evidenced by scientific papers from these labs published in Science in January 2013. Indeed, the brief argues that Dr. Zhang's alleged reduction to practice in June 2012, after receiving the information regarding sgRNA from Dr. Marraffini, is itself evidence of CVC's complete conception, because the information Dr. Marraffini communicated to Dr. Zhang came from Jennifer Doudna and Emmanuelle Charpentier. The brief provides a convenient checklist of that disclosure:
CVC's derivation argument rests crucially on Dr. Marraffini's testimony. As set forth in the brief, this deposition testimony established derivation of the Broad invention from CVC's inventors:
Before he attended the 2012 CRISPR Conference, Marraffini had already reviewed CVC's draft Jinek 2012 manuscript as one of the peer-review referees for Science. CVC submitted the draft Jinek 2012 manuscript to Science by June 8, 2012, just before the conference. The draft Jinek 2012 manuscript revealed to Marraffini that processed tracrRNA, mature crRNA, and Cas9 are the necessary and sufficient components of the catalytic CRISPR-Cas9 DNA-cleavage complex, the design and use of a functional sgRNA CRISPR-Cas9 system, and also expressly stated that the sgRNA CRISPR-Cas9 system could be used for "genome editing in cells of the three kingdoms of life for biotechnological, biomedical and gene therapeutic purposes."
Marraffini recalls that CVC's disclosure of tracrRNA as a necessary component in the catalytic DNA cleavage complex "as well [as] the development of the [sgRNA]" were "the most important findings that the team made" because, before CVC's breakthrough, no one knew that tracrRNA was an active component in the complex. Before June 26, 2012, Zhang had no concept of a sgRNA CRISPR-Cas9 system of Count because he was unaware that tracrRNA was a necessary component of the catalytic CRISPR-Cas9 DNA-cleavage complex. . . . Prior to June 2012, the art taught that tracrRNA was merely an intermediate cofactor that RNase III and Cas9 needed for RNA processing to generate mature crRNA guides from long, unprocessed pre-crRNAs. Marraffini confirmed [this understanding in the art], testifying that tracrRNA was only "known . . . to be involved in the generation of guide RNAs." Marraffini also testified that, before June 26, 2012, he and Zhang only discussed "the importance of tracr for what we knew at the time was generation of RNA guides," and that "the system, as we knew it, required RNase III to generate guides." [emphasis in brief]
Marraffini explained further that he remembered this discovery as "the most important findings that the team made, the fact that the tracrRNA is required for cleavage of DNA, which was not known at the time. And I thought that was a very important finding." Marraffini's review [of the Jinek manuscript] showed that he understood the CVC inventors intended to use their sgRNA CRISPR-Cas9 system in eukaryotic cells: Being guided by the crRNA sequence, the dsDNA nuclease activity of Cas9 could be re-programmed to target essentially any sequence in any genome with a very high specificity. Perhaps in one of the most exciting experiments of this work, Jinek et al. show that Cas9 can be 'loaded' with synthetic crRNAs that direct any chosen sequence in a plasmid. As the authors indicate, this provides a new, invaluable tool for genome editing. Marraffini's [contemporaneous] reviewer comments comport with his deposition testimony that CVC's sgRNA was "a fundamental advance that we could use in our project." After reading the published version of Jinek 2012 in Science, Zhang similarly stated in a June 30, 2012 email to Marraffini that Jinek 2012 was "a great paper" and made Zhang feel that CVC was "moving in [the] direction" of eukaryotes because the abstract refers to "genome editing" and the paper tested eukaryotic target gene (GFP) sequences.
Like the other attendees at the June 21 CRISPR Conference, Marraffini knew that he and his collaborator Zhang were now in a race to be the first to apply CVC's sgRNA CRISPR-Cas9 system for gene editing in eukaryotic cells and publish the results. After returning from the conference, Marraffini promptly emailed Zhang on June 26,[2012], requesting a phone call to discuss "a couple of 1 presentations" he saw at the conference that were "important" to their project. Marraffini clarified on cross-exam that the important presentation he had in mind for "gene editing of human cells" was indeed "the presentation by . . . Jinek and Chylinski about the single-guide RNA." Zhang quickly responded to Marraffini's email, and the two immediately set up a phone call to discuss the CVC presentation. On that call, he and Zhang discussed CVC's sgRNA CRISPR-Cas9 system and how CVC's sgRNA was a "fundamental advance that we could use in our project." In particular, Marraffini testified that he "must have" told Zhang about the role of tracrRNA in the catalytic DNA-cleavage complex. (Marraffini testifying that prior to June 26, his conversations with Zhang were "only about the importance of tracr for what we knew at the time was generation of RNA guides.") Marraffini also communicated to Zhang the intended purpose of using CVC's sgRNA CRISPR-Cas9 system in eukaryotic cells:
Q: So you . . . did you generally convey the view to Dr. Zhang that you thought the single-guide RNA would be an important tool for genome editing?
A: Yeah. Yes, I -- that's why I sent him the information, yes.
When asked if he communicated to Zhang that CVC's sgRNA CRISPR-Cas9 system would be an important tool for genome editing in eukaryotes specifically, Marraffini stated unequivocally, "yes":
Q : [W]hen you first conveyed to Dr. Zhang the single -- single-guide RNA system described in Jinek 2012, did you convey to him that you thought it would be an important tool for genome editing in eukaryotes specifically?
A: Yes.
Marraffini also disclosed to Zhang that there was no need to employ the cumbersome RNA-processing system with which Zhang had been unsuccessfully tinkering. After their June 26 phone conversation, Marraffini sent a follow-up email to Zhang, stating that he "checked" the Jinek 2012 draft manuscript and "[CVC] provide pre-processed crRNAs to the in vitro reaction, so there is no need for RNAse III . . . ." Marraffini testified that until that time, "the system, as [he and Zhang] knew it, required RNase III to generate guides." Indeed, prior to June 26, all of Zhang's CRISPR experiments were failed attempts to mimic pre-crRNA processing, evident from his repeated use of unprocessed crRNAs, unprocessed tracrRNA, and RNase III in the experiments When asked on cross-exam whether he had ever used pre-processed (mature) crRNA or processed tracrRNA in his experiments before June 26, Zhang testified that "I don't have a clear recollection if I have done any experiments [with mature crRNA]" and "I don't remember if I had done experiment there, with that [processed tracrRNA]."
Zhang admits in his declaration that he obtained the sgRNA design from Marraffini, although he conspicuously omits the fact that the "chimeric RNA" Marraffini shared with him was CVC's sgRNA. At deposition, however, Zhang conceded that the sgRNA structure indeed came from "the Doudna-Charpentier group." Marraffini—who provides third party testimony that was unchallenged by Broad at deposition—testified that the sgRNA image in his June 26 email was a direct copy from the then-unpublished Jinek 2012 manuscript he had reviewed:
Q: Where did you get that particular image from, Doctor?
A: From the [Jinek 2012] manuscript. But it was the same one that it was public from the conference.
The sgRNA structure Marraffini sent to Zhang in his June 26 email is identical to the "chimera A" sgRNA structure [set forth above herein] Jinek and Chylinski presented on June 21, which is also identical to the sgRNA Zhang claims he "designed" on June 27, 2012 [all references to exhibits omitted].
Based on this testimony, CVC argues that the evidence shows that Broad's inventors, and specifically Dr. Zhang, derived their claimed invention from the CVC inventors through Dr. Marraffini. The brief further characterizes (turnabout being fair play) their own litany of purported "failures" by the Zhang lab, particularly with regard to the "fallback position" set forth in Broad's priority brief, that work performed prior to CVC's March 2012 conception date should be considered by the Board in making its priority determination. This work, on alternative systems for gene editing (e.g., TALEN and ZFN), is not CRISPR, CVC argues, and without knowledge of CVC's sgRNA discovery the Broad inventors were pursuing (fruitlessly) more complicated versions of CRISPR that required, inter alia, RNAse III and unprocessed tracr and crRNA fragments. Indeed, CVC argues that until Dr. Marraffini's disclosure of its invention to Dr. Zhang there was no recognition by the Broad inventors of the role and importance of tracr RNA.
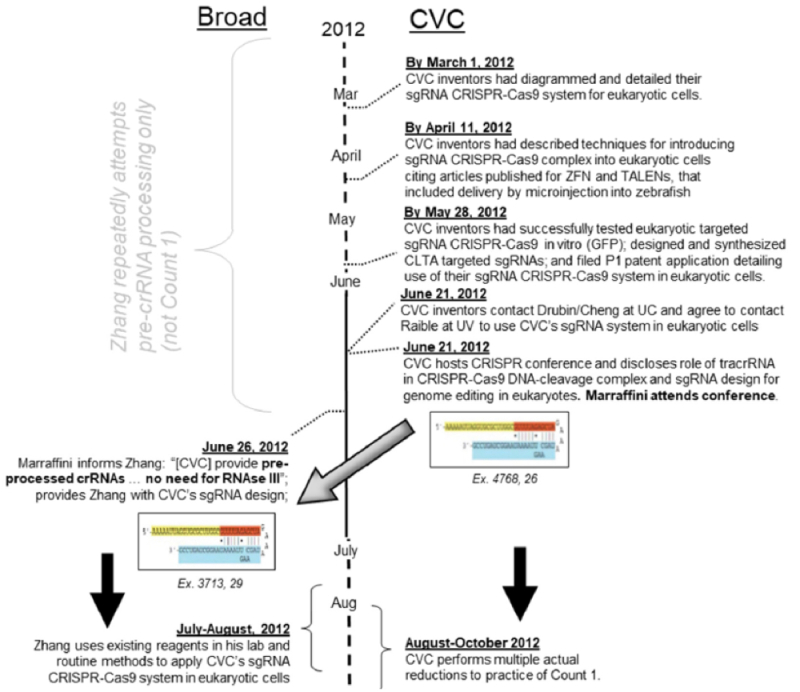
CVC sets out its own summary timeline of events by the parties, modifying the timeline presented in Broad's priority motion according to CVC's version of events as supported by the evidence set forth in this brief:
With regard to their purported incomplete conception argued by Broad, CVC argues that it had recognized and addressed each and every one of these putative deficiencies. In addition, CVC argues that each of these "adaptations" required to practice CRISPR in eukaryotic cells "is not an invention, is not in the count, and is not required for reduction to practice," providing a mantra repeated for each of the various adaptations asserted by Broad:
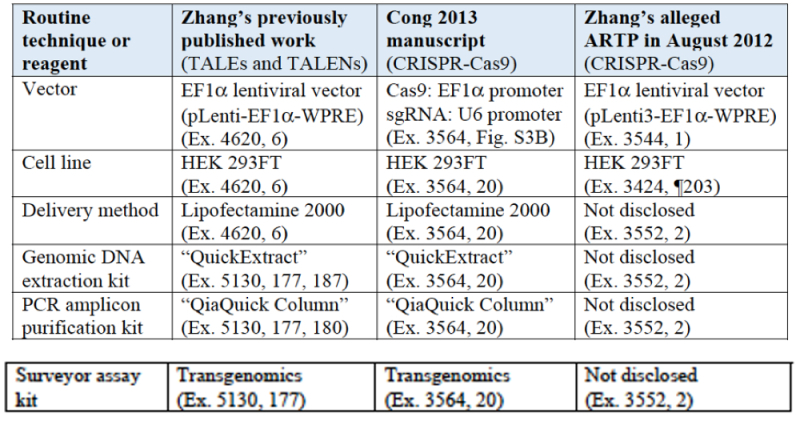
(Missing, of course because it belongs properly in CVC's reply to Broad's opposition to their priority motion, is an explanation of the litany of CVC's failures recited in Broad's opposition.)
Turning to Broad's evidence of actual reduction to practice, CVC makes the argument that the evidence is uncorroborated, because three of the witnesses (Sanjana, Cong, and Cox) set forth in this table are named as inventors, citing Chen v. Bouchard, 347 F.3d 1299, 1309 (Fed. Cir. 2003):
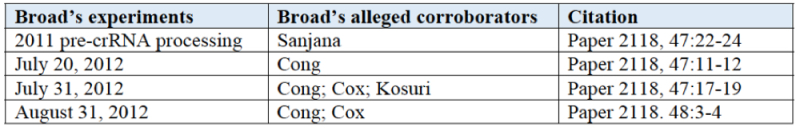
And the fourth, Kosuri, does not corroborate Dr. Zhang's testimony regarding ARTP, relying on vague ("this system") contemporaneous representations from Dr. Zhang and not having firsthand knowledge.
The brief further alleges failure of proof, no contemporaneous inventor recognition, and failure of corroboration, for each of Broad's purported actual reductions to practice of eukaryotic CRISPR on July 20, July 28, July 31, and August, 2012, and further argues that because Broad did not establish the actual August date for this last instance of ARTP, the Board should consider it to be August 31, which is after CVC's August 9, 2012 asserted ARTP date.
Broad's Reply brief to this opposition is due April 6, 2021.