On December 3rd, Senior Party Sigma-Aldrich filed its Substantive Preliminary Motion No. 1 in Interference No. 106,133 (which names the Broad Institute, Harvard University, and MIT (collectively, Broad) as Junior Party), asking the Patent Trial and Appeal Board to deny Broad benefit of its U.S. Provisional Application No. 61/736,527, filed December 12, 2012 (termed "P1"), pursuant to 37 C.F.R. § 41.121(a)(1) and S.O. ¶¶ 121 and 208.4.2. The basis for Sigma-Aldrich's motion is that this application does not disclose a constructive reduction to practice of an embodiment within the scope of Count 1 of the interference as declared. Sigma-Aldrich specifically contends that the P1 application does not show either than Broad's inventors had achieved a CRISPR-Cas9 system that cleaved a targeted DNA molecule and altered the expression of the gene product of that cleaved molecule, nor did the P1 application disclose introducing Cas9 protein into a eukaryotic cell with only one linked nuclear localization signal ("NLS") that resulted in CRISPR-mediated gene editing (Sigma-Aldrich asserts that all of the P1 disclosure involved Cas9 modified with 2 NLS sequences).
After setting out the conventional requirements for priority benefit with regard to satisfying the written description requirement of 35 U.S.C. § 112(a), and the qualifications and characteristics of a person of ordinary skill in the art in December 2012, the brief highlights certain portions of the Count Sigma-Aldrich argue are not adequately disclosed in Broad's P1 application:

Sigma-Aldrich argues that Broad merely identified the existence of CRISPR-mediated DNA cleavage and did not establish resulting changes in gene expression. Parsing the P1 disclosure, Sigma-Aldrich argues that the P1 specification itself sets forth definitions regarding the meaning of altering gene expression as recited in the Count, and distinguishes this from modifying the sequence of a targeted gene, quoting the specification as reciting in the alternative: " a modification in one or more nucleic acid sequences associated with a disease, or an animal or cell in which the expression of one or more nucleic acid sequences associated with a disease are altered." These distinctions are supported by portions of the P1 specification that describe embodiments where a target polynucleotide is altered by introduction of a heterologous sequence, and other embodiments where expression of a targeted sequence is modified by increasing or decreasing its expression (i.e., without necessarily altering the targeted gene's expression). Sigma-Aldrich further asserts that these distinctions are supported by P1 disclosure relating to the assays used to detect changes in expression that differ from assays used to detect changes in the sequence. Sigma-Aldrich then argues that Broad's P1 application reports no "direct analyses of expression of any targeted gene product," whether with or without CRISPR-mediated editing. And the only data presented in the P1 application relating to gene expression (Figures 1B, 6, and 13) was concerned with expression of the components of the CRISPR-Cas9 gene editing system (SpCas9 and RNase III). Nor would the skilled worker have apprehended any demonstration of using CRISPR in a eukaryotic cell to alter gene expression:
• "[non-homologous end joining (]NHEJ repair of Cas9-mediated cleavage created some nucleotide insertions/deletions (or "indels") to the emx1, pvalb, and th genes, which were detected indirectly by a Surveyor assay;"
• "deletion of a 118-bp sequence of a human emx1 gene as a result of cleaving two target sites (albeit noting that "it would be impossible to predict the nature of the final mRNA or protein that could have been produced, or whether the expression of the mRNA or EMX1 protein had been altered at all, e.g., whether the expression of the gene product had increased or decreased);" and
• "one example of gene editing where a 12-bp sequence comprising two restriction enzyme sites replaced the stop codon in the emx1 gene [but the data would have only established the ("minor") change in sequence but not necessarily expression, for which there was no experimental disclosure]."
Sigma-Aldrich further argued that in these instances there was no experimental data in the P1 provisional application disclosing changes in the level of expression in, inter alia, the emx1 gene (Sigma-Aldrich illustrates the principle that altering sequence may or may not alter expression with regarding to producing a library of expressed proteins in yeast comprising C-terminal green fluorescent protein fusions, in which addition of this tag did not significantly alter gene expression.)
Sigma-Aldrich also argued that the Broad P1 priority document did not disclose Cas9 proteins altered by introduction of only a single NLS sequence, as recited in the Count:

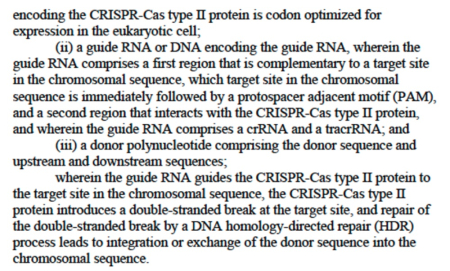
Not only does the P1 provisional application not disclose Cas9 altered by introduction of only a single NLS, Sigma-Aldrich argued that the specification described failure to achieve CRISPR cleavage in a eukaryotic cell using such single NLS embodiments of Cas9. Consequently Sigma-Aldrich notes that "all of the examples disclosed in Broad P1 use two NLSs for targeting Cas9 to the eukaryotic cell's nucleus for gene editing." Sigma-Aldrich emphasized the P1 disclosure of experiments investigating nuclear localization of three SpCas9 constructs, wherein:
[E]ach [was] expressed as a fusion protein with the florescent protein marker, GFP, at their C-terminus, and with: (1) an NLS attached to the N-terminus of SpCas9 (i.e., NLS-SpCas9-GFP); (2) an NLS attached to the C-terminus of the GFP (i.e., SpCas9-GFP-NLS); and (3) an NLS attached to both N- and C termini of SpCas9-GFP (i.e., NLS-SpCas9-GFP-NLS, which was referred to as 2xNLS12SpCas9)
In these experiments, Sigma-Aldrich argued, only species (3) (having 2 NLS sequences) were successful at achieving CRISPR cleavage in a eukaryotic cell. This conclusion was supported in the brief by illustrations that "[a]ll CRISPR-Cas gene editing examples disclosed in Broad P1 use[d] two NLSs:
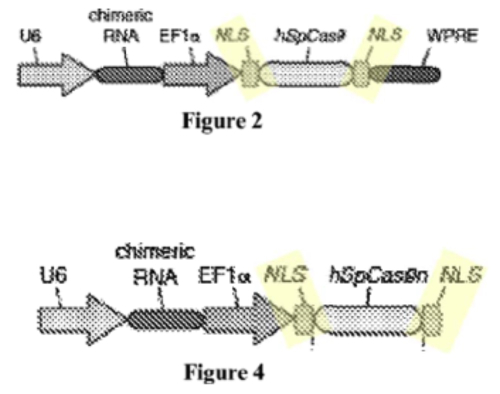
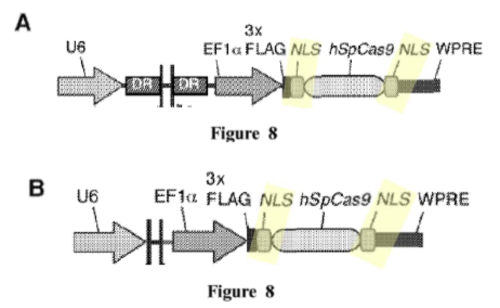
And that only constructs having 2 NLSs were functional:
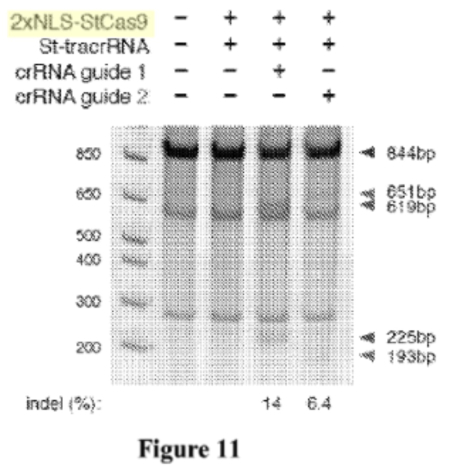
Sigma-Aldrich concedes that Broad's P1 application contains disclosure related to single NLS-bearing Cas9 proteins. Sigma-Aldrich terms this disclosure "speculative," due to the absence of any experimental demonstrations showing successful CRISPR-Cas9 cleavage using single NLS-bearing Cas9 species.
Unlike conventional Preliminary Motion practice as Senior Party Sigma-Aldrich does not need the Board to grant this motion to change any evidentiary burdens, but success would increase the time between Sigma-Aldrich's earliest priority date and its own, which could have its own advantages.