
Ulrich (Uli) Laemmli, an illustrious professor of biochemistry and molecular biology, developer of SDS (sodium dodecyl sulfate)-polyacrylamide electrophoresis (PAGE) for separating proteins, and responsible for identifying the "scaffold" structure of human chromosomes, was fond of saying (when he was a professor at Princeton) that in biological research it was helpful (if not essential) to "look for the mutant." Difficulties in following this advice productively include the (low) frequency with which mutants arise, the fact that most such mutants lose or compromise the function(s) of the encoded proteins, and that conventional methods for producing mutants are as likely as not to be lethal to cells (either due to mutants of the protein of interest or other proteins that are mutated in passing). So-called "targeted" mutations produced, in its most au courant version, by CRISPR-Cas9 (wherein CRISPR is an acronym for Clustered Regularly lnterspaced Short Palindromic Repeats) require knowledge of the site to be mutagenized and hence comprise only a limited subset of targets for productive mutagenesis.

A group of researchers* from Berkeley have used a modified version of Cas9 (nCas9, bearing a D10A mutation that produces "nicks" in target DNA) to produce a system they term EvolvR for dramatically (7,700,000-fold) increasing in vivo mutagenesis, as reported in the scientific journal Nature in a Letter entitled "CRISPR-guided DNA polymerases enable diversification of all nucleotides in a tunable window." This is illustrated in E. coli bacteria for producing cells resistant to the antibiotic streptomycin. The modified nCas9 is fused to the amino terminus of a fidelity-reduced variant (D424A, I709N, A759R) of E. coli DNA polymerase I. The plasmid encoding this construct also encoded a guide DNA (gDNA) to target a second plasmid containing a homologous gene sequence. These model experiments showed mutations (of all expected types) arising within a 17 nucleotide window 3' of the nick site, consistent according to these authors with the known 15-20 nucleotide processivity of the polymerase, as illustrated below. There were also a low frequency of 5' mutations that the authors speculated may have been due to the polymerase's 3'-5' exonuclease activity. Also tested was a reversion assay, wherein the target plasmid encoded a streptomycin resistance gene bearing a nonsense mutation. Against a measured background mutation rate of 10-10 mutations/nucleotide/ generation, consistent with E. coli mutation rates, introducing the EvolvR plasmid into these cells "markedly increased the mutation rate at the targeted locus 24,500-fold over the wild type while increasing the global mutation rate 120-fold over the wild type," which the authors state is similar to other mutagenesis methods used in E. coli.
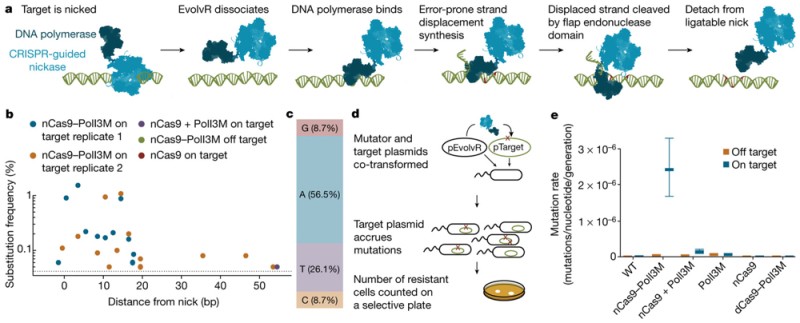
Mutation rates using this system were further enhanced by making additional mutations in nCas-9 (K848A, K1003A, R1060A); these mutations had been suggested in the art to "lower the non-specific DNA affinity of Cas-9." These plasmids "increased the global mutation rate 223-fold compared to wild-type cells (1.9–fold greater than nCas9–PolI3M), yet elevated the mutation rate at the targeted locus by 212,000-fold (8.7-fold greater than nCas9–PolI3M)." The mutagenesis rate was further increased to the maximum reported (7,700,000-fold) by further modifications (D424, I709N, F742Y, A759R, and P796H) to the polymerase portion of the fusion protein. Alternative polymerases (from the bacteriophage Phi129) did not achieve this high a mutation rate, nor was inserting the thioreductase-binding domain of bacteriophage T7 DNA polymerase into the PolI sequence as effective (although this change did increase the reach of mutation rate in the target DNA). Other refinements included removing internal ribosome binding sites in the PolI sequence to reduce (4.14-fold) "off-target" mutagenesis.
The authors also report the ability to target separate DNA target sequences, although they note that when those targets are within 110 bp of each other, the nicking activity should be directed to the same strand to avoid lethality.
The paper ends with the following hopes for future applications of the technology:
EvolvR offers the first example of continuous targeted diversification of all nucleotides at user-defined loci, which will be useful for evolving protein structure and function, mapping protein–protein and protein–drug interactions, investigating the non-coding genome, engineering industrially relevant microbes and tracking the lineage of cell populations that cannot tolerate double-stranded breaks. As a guiding principle for using this tool, our data suggest that 1µl saturated E. coli culture expressing enCas9–PolI3M–TBD for 16h contains all single substitutions in the 60-nucleotide window with more than tenfold coverage. Future work towards adapting EvolvR for use in cells possessing low transformation efficiency, as well as increasing the mutation rate and window length of EvolvR mutagenesis, would enable new forward genetic applications.
*Shakked O. Halperin, Connor J. Tou, Eric B. Wong, Cyrus Modavi, David V. Schaffer & John E. Dueber