Genetic instability has long been recognized as a hallmark of oncogenesis and tumor progression. The phenomenon was first identify cytogenetically, most famously by the Philadelphia chromosome in chronic myelogenous leukemia (see Wapner, The Philadelphia Chromosome: A Genetic Mystery, a Lethal Cancer, and the Improbable Invention of a Lifesaving Treatment), later appreciated as the result of a translocation between a human protooncogene (c-abl) with a heterologous sequence that resulted in development of the disease. But it was later recognized by histological analysts that cancer cells could be characterized by a host of aneuploidies, including chromosome loss and duplication as well as changes in fine structure features of the genome.
The advent of rapid and (relatively) inexpensive whole genome sequencing (WGS) methodologies has resulted in even more sensitive assessments of changes in human genomic DNA associated with cancer progression. This week, the scientific journal Nature published a paper* entitled "Pervasive lesion segregation shapes cancer genome evolution" that showed for the first time a heritable pattern of DNA strand-specific changes caused by contact with chemical mutagenic agents and the consequences for cancer development. The experiments were performed by treating inbred male C3H/HeOuJ mice (as well as "divergent" CAST/EiJ mice) with one dose of dimethynitrosamine (DEN), a known cancer-causing agent. Treatment with this mutagenic agent were found to be predominantly (76%) mutations in A:T basepairs (T→N or A→N, where N is any of the other bases), which the paper note is "consistent with the long-lived thymine adduct O4-ethyl-deoxythymidine being the principal mutagenic lesion." Whole genome sequencing (WGS) was performed on a total of 371 independently-arising tumors from 104 C3H mice and these researchers reported finding about 60,000 point mutations in each tumor (about 13 changes per megabasepair, Mb). Perhaps unsurprisingly (in view of what is known about how this chemical produces mutations), insertion and deletion mutations were rarely detected.
Upon analysis of these mutations, the authors found "multimegabase genomic segments with pronounced Watson-versus-Crick-strand asymmetry of mutations, frequently encompassing entire chromosomes" (i.e., "an excess of T→N over A→N mutations when called on the forward strand of the reference genome, and Crick-strand bias as the converse of this"), with s median span of these lesions being 55Mb long. The researchers state that this outcome "DEN-induced lesions remaining unrepaired before genome replication." The mechanistic consequence is shown in Figure 2i:
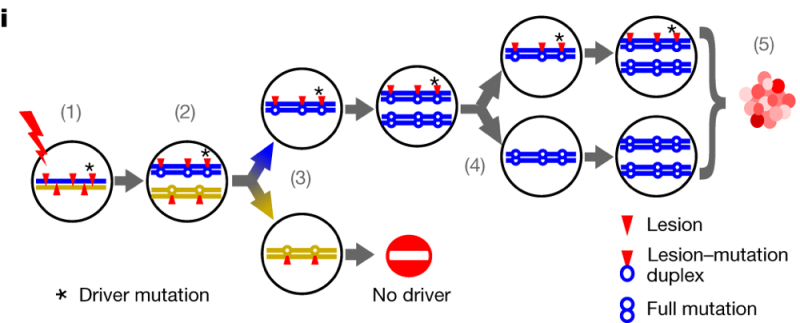
where "(1) A mutagen generates lesions (red triangles) on both DNA strands. (2) If not removed, lesions will segregate into sister chromatids: one carrying only Watson-strand lesions (blue) and the second carrying only Crick-strand lesions (gold). Postmitotic daughter cells will have independent lesions and resulting replication errors (3), resolved into full mutations in later replication (4). (5) Only lineages containing driver changes (* in (1)) will expand into substantial populations."
As the authors explain, "[a]symmetric regions show a 23-fold excess (median) of their preferred mutation over its reverse complement, thus more than 95% of lesions that generate a mutation segregate for at least one mitotic division," a phenomenon they term "lesion segregation." The authors explain instances where these asymmetries change in a chromosome as being the result of sister chromatid exchanges resulting from homologous recombination-mediated DNA repair events. While these asymmetries are reported to be equally distributed on the Watson and Crick strands in the genome, this is not observed at loci encoding genes associated with tumorigenesis (what the researchers term "driver genes": Braf, Hras, and Egfr). These sites show "oncogenic selection," defined as a bias for retaining mutations associated with their role in oncogenesis. The researchers also found transcription-coupled nucleotide-excision repair of such lesions preferentially in portions of the chromosome encoding the template (mRNA) strand of genes in the chromosomes, wherein "mutations in highly expressed genes were reduced by 79.8 ± 1.0% (mean ± s.d.) if the tumour had template-strand lesions."
Discovery of lesion segregation provides a mechanism for the genetic heterogeneity found in many tumors. As the authors explain, "[a] segregating lesion may act as template for multiple rounds of replication in successive cell cycles (as shown in the Figure). Each replication could incorporate different incorrectly or correctly paired nucleotides opposite a persistent lesion, resulting in multiple alleles at the same position. Consistent with this notion, multiallelic mutations have been reported in human cancers . . . ." Here, 8% of mutated sites showed multiallelic variants (amounting to 1.8 million sites in C3H tumors induced by DEN), whereas only 0098% of sites between tumors showed nucleotide changes. The consequence: "[t]he generation of multiallelic variation produces combinatorial generic diversity that would not be expected under purely clonal expansion." The researchers explain the consequence of these results:
Tumours with high rates of genetic diversity have consistently high rates of multiallelism throughout their genome[]. They are likely to have expanded from a first-generation daughter of the original DEN-mutagenized cell, in which all DNA is a duplex of a lesion-containing and non-lesion-containing strand. Therefore, replication using lesion-containing strands as the template in subsequent generations produces multiallelic variation uniformly across the genome. Tumours with lower total levels of genetic diversity exhibit discrete genomic segments of high and low multiallelism[]. These tumours probably developed from a cell some generations after DEN treatment. Each mitosis following DEN exposure is expected to dilute the number of lesion-containing strands in each daughter cell by approximately 50%. Only lesion-retaining fractions of the genome generate multiallelic and combinatorial genetic diversity in the daughter lineages; consistent with this, the multiallelic segments mirror the mutational asymmetry segmentation pattern.
The consequences of DEN mutagenesis reported are striking: "[i]n 67% of C3H tumours and 21% of CAST tumours, the initial burst of mutations was instantly transformative."
Lesion segregation can also be observed, as reported in this paper (if you know to look for it) in chemical mutagenized human induced pluripotent stem cells (iPSCs), as well as in tumors induced by sunlight (UV irradiation), tobacco smoke (benzo[a]pyrene diol-epoxide) and certain chemotherapeutic agents. The authors note, however, that unlike experimentally induced tumors most naturally occurring tumors are the result of a series of longitudinal genetic insults rather than a single injury. Accordingly, they expect that lesion segregation may be masked as a consequence of these multiple insults. Nevertheless, they also report finding lesion segregation in renal, biliary, and hepatic tumors from a human cancer genome compendium (n=18,850 tumors from 22 primary tumor sites).
The paper summarizes the significance of their findings thusly:
Once identified, lesion segregation is a deeply intuitive concept. Its practical applications provide new vistas for the exploration of genome maintenance and fundamental molecular biology. The discovery of pervasive lesion segregation profoundly revises our understanding of how the architecture of DNA repair and clonal proliferation can conspire to shape the cancer genome.
* Sarah J. Aitken, Craig J. Anderson, Frances Connor, Oriol Pich, Vasavi Sundaram, Christine Feig, Tim F. Rayner, Margus Lukk, Stuart Aitken, Juliet Luft, Elissavet Kentepozidou, Claudia Arnedo-Pac, Sjoerd V. Beentjes, Susan E. Davies, Ruben M. Drews, Ailith Ewing, Vera B. Kaiser, Ava Khamseh, Erika López-Arribillaga, Aisling M. Redmond, Javier Santoyo-Lopez, Inés Sentís, Lana Talmane, Andrew D. Yates, Liver Cancer Evolution Consortium, Colin A. Semple, Núria López-Bigas, Paul Flicek, Duncan T. Odom & Martin S. Taylor.
Cancer Research UK Cambridge Institute, Department of Pathology, University of Cambridge, UK; Department of Histopathology, Cambridge University Hospitals NHS Foundation Trust, UK; MRC Human Genetics Unit, MRC Institute of Genetics and Molecular Medicine, School of Mathematics and Maxwell Institute, Higgs Centre for Theoretical Physics, University of Edinburgh, UK; Institute for Research in Biomedicine (IRB Barcelona), The Barcelona Institute of Science and Technology, Barcelona, Spain; European Molecular Biology Laboratory, European Bioinformatics Institute, Hinxton, UK; Edinburgh Genomics (Clinical), The University of Edinburgh, Edinburgh, UK; Universitat Pompeu Fabra (UPF), Barcelona, Spain; Institució Catalana de Recerca i Estudis Avançats (ICREA), Barcelona, Spain; German Cancer Research Center (DKFZ), Division of Regulatory Genomics and Cancer Evolution, Heidelberg, Germany.